General Questions
1. What is FlyExpress? How do I get started using FlyExpress?
FlyExpress is an online platform for biological discovery. FlyExpress contains a digital library of standardized images from BDGP, Fly-FISH and peer-reviewed publications capturing the spatial expression of thousands of genes at different developmental stages in the fruit fly Drosophila melanogaster. An image-matching search engine and genome-wide summaries of spatiotemporal expression patterns (GEMs) reveal genes with similar or overlapping patterns of expression in the developing embryo. Motif-finding tools integrate image and sequence data to identify genomic elements involved in regulation in upstream regions.
2. Are there other websites that can teach me more about Drosophila?
Yes, there are a number of internet resources containing vast amounts of information on Drosophila, including:
- FlyBase - http://www.flybase.org
- FlyMine - http://www.flymine.org
- BDGP - http://www.fruitfly.org/
- Fly-FISH - http://fly-fish.ccbr.utoronto.ca/
- The Interactive Fly - http://www.sdbonline.org/fly/aimain/1aahome.htm
- FlyAtlas - http://www.flyatlas.org/
- FlyMove - http://flymove.uni-muenster.de/
In addition, the following link contains relevant web-based databases of Drosophila genomics: http://flybase.org/static_pages/allied-data/external_resources5.html
3. Which is the best web browser to use for FlyExpress?
FlyExpress is designed to work on all web browsers. Please let us know if you have difficulty in using the FlyExpress website on any particular operating system or web explorer.
4. If I find dead links, bugs, an image that is classified incorrectly, or an inaccurate spatial profile, where can I report it?
We have thorough quality control procedures to ensure that images are classified properly. However, errors may still arise. Please inform us of any errors you encounter through our Feedback page. Provide the image identifier and indicate what you think is wrong with the picture or its annotation.
5. How is FlyExpress useful to my research?
FlyExpress is useful to: (a) view the expression time-course of genes throughout development at a cellular level, (b) find co-expressed genes during a particular developmental stage range, (c) analyze gene lists for enrichment of function and involvement in gene interactions, (d) find publications containing images with co-expressed genes, and (e) view global genome-scale expression profiles at different stages of development. Newly developed tools in FX7 allow for the: (f) pairwise similarity analyses of upstream intergenic regions (IGRs) via a dotplot viewer .
6. What is a GEM?
Genome-wide Expression Maps are summaries of spatial patterns of all genes expressed at a given stage. GEMs are shown as heat maps, with the darkest regions indicating domains where the largest numbers of genes are expressed.
Image Data & Annotation
7. How are in situ images annotated?
Our current image annotations - including developmental stage ranges, gene/EST probed, and the controlled vocabulary terms - come directly from BDGP or Fly-FISH. In Fly-FISH original images containing multiple embryos, we assigned stage ranges to any embryos lacking that information. In peer-reviewed publications, images were annotated through a combination of FlyBase-Harvard curators and our in-house staff. We have standardized all the images for uniformity and computational analysis, and have also annotated the anatomical orientation (e.g., lateral, dorsal, ventral).
8. Some embryo images show no gene expression when there actually is expression during a particular stage. Also, some images have squashed embryos. Why are these included in FlyExpress?
There are very few low-quality images in BDGP and Fly-FISH datasets, and we are aware that such issues are inevitable in high-throughput in situ hybridization experiments. However, we incorporate all images of full embryos into FlyExpress, as we do not, by policy, delete published data from our collection.
9. I looked at the in situ images for a gene I study, and some of them show only artifact staining. Why are these included in FlyExpress?
Artifact staining of some embryos, especially those in the later stages of embryogenesis, is an inevitable consequence of high-throughput in situ hybridization. However, we incorporate all images of full embryos into FlyExpress, as we do not, by policy, delete published data from our collection.
10. Why are there stage 17 embryos in the BDGP stage 13-16 category?
BDGP uses the 16-stage system, not the 17-stage system. Many landmarks of embryogenesis are the same in both staging systems, but there are some differences. We, by policy, faithfully transmit the information provided by the original authors of the images.
For further information about the 16-stage system, refer to: Roberts, D.B., ed. Drosophila: A Practical Approach. USA: Oxford University Press, 1998. For further information about the 17-stage system: http://www.sdbonline.org/fly/aimain/2stages.htm.
11. Where can I obtain information about protocols and specific riboprobes used?
The BDGP website provides their detailed in situ protocol and probe information at their website, http://www.fruitfly.org. Fly-FISH also provides detailed information at their website, http://fly-fish.ccbr.utoronto.ca/.
12. What alleles for each gene are in FlyExpress?
All BDGP and Fly-FISH images show in situ expression patterns for the wild-type allele of each gene. Alleles for publications depend on the specific paper.
Searching for Co-Expressed Genes
13. Why has FlyExpress replaced pairwise with GEM-based searching?
Genome-wide Expression Map (GEM)-based searching is less computationally intensive, and eliminates the need to pre-compute an enormous pairwise image similarity matrix that contains hundreds of millions of values and requires a large-scale database indexing system. GEM-based searching provides almost instantaneous result returns, impressive for a database of over 100,000 high-resolution images.
14. What is the difference between pairwise and GEM-based searching?
To calculate the pairwise similarity between two black-and-white expression profiles, the intersection of black pixels is divided by the union of black pixels. In GEM-based searching, selecting from a finite number of normalized expression profiles produces a set of unique search pixels representing key points of gene expression. Co-expressed genes are found by taking the collection of pixels in a spatial profile, mapping them onto the appropriate GEM, and asking for genes that share the highest numbers of co-expressed pixels.
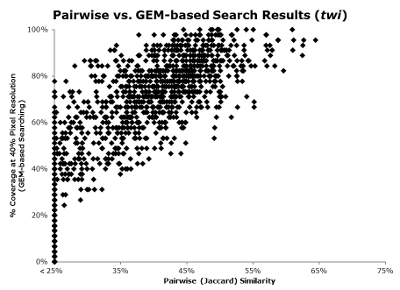
15. Do pairwise and GEM-based searching produce similar results?
Yes. A comparison of search results for the same query of a stage 7-8 lateral image depicting the expression of twist (FBim9035159_c), reveals a strong correlation, below (click for a larger view):
16. What are the black and white images (spatial profiles) next to the color images?
FlyExpress utilizes a normalization protocol that generates images of standard size (320x128 pixels) to facilitate comparisons between images. The standardized color images are converted to binary black & white using a threshold algorithm. The image standardization procedure is described in Konikoff et al. 2012.
17. Why are there three spatial profiles to choose from?
A major functionality of FlyExpress is the "Find Similar Patterns" feature, which rapidly searches the Digital Library to find co-expressed genes. The "Select a black-and-white spatial profile and pixel sampling level to customize your search" option maximizes the search customization by allowing for three different thresholds for each standardized image.
18. What are some disadvantages of relying heavily upon computational methods to analyze gene expression similarities and differences that I should be aware of?
While FlyExpress provides a convenient method of finding co-expressed genes, expression pattern similarity may be overestimated or underestimated for numerous reasons. For example, computers cannot decipher between legitimate expression and noise. If two genes have the same expression pattern, but one gene has images with artifact staining while the other does not, expression similarity will be underestimated. So, we recommend that the results produced be used as a guide, rather than the final word on the genes with overlapping patterns of expression.
Pairwise Gene Comparisons
19. How can I compare upstream sequences of two co-expressed genes?
Click the "View" button in the leftmost column of your co-expression search results to compare the selected gene with the query. Alternatively, type in the following URL to access any pairwise comparison you wish to make on this page directly: http://www.flyexpress.net/search/pairwise/GeneA/GeneB (replace GeneA and GeneB with the genes' symbols).
20. What are the data sources for the information displayed on this page?
Data displayed on the page comes from the following sources:
- Genome/Track Data - The latest version of FlyBase
- TFBS - REDfly
- Conservation Scores - Averaged PhastCons 27-way values, via UCSC Genome Browser
- Dotplot IGR Sequences – The latest release of FlyMine
21. What color scale is used on the dotplot when the Evolutionary Filter is selected?
To produce each dotplot point, PhastCons 27-way scores were averaged and color-coded according to evolutionary conservation level. Highly conserved regions are displayed as black dots, with lighter colors indicating less conservation. When no evolutionary data is available for a specific region of the genome, no color-coded stretches of k-mers are displayed in the dotplot.
PhastCons Average Dotplot Color 0-0.25 White >0.25 - 0.5 Light Gray >0.50 - 0.75 Dark Gray >0.75-1 Black
Citation/Copyright Info
22. Can I download images?
While images are not encrypted or otherwise digitally protected, some images in the extensive FlyExpress image database are copyright-protected. Therefore, permission should first be obtained from the relevant copyright owner. Please see the Copyright notice that appears on the Home page.
23. What is the relevant copyright information pertaining to GEMs and dotplot images? Can I include them in my manuscript?
You are welcome to use GEMs and dotplot images in your publications. We request that you cite the latest FlyExpress resource paper whenever you use it for research purposes. FlyExpress reference information is provided on the front page.
24. How do I cite FlyExpress Motif Discovery in my manuscript?
Please cite the latest FlyExpress resource.